Introduction
Recent decades have seen major demographic changes in the Arctic, with Indigenous populations undergoing a transition from a nomadic mode of life to living in more densely populated communities. At the same time, current trends in climate change are resulting in a significantly warmer global environment, with a dramatic impact on the Arctic that is already beginning to be manifested. In industrialized regions the incidence of infections with foodborne pathogenic bacteria tends to vary with the seasons, with the warmer seasons being more propitious for the spread of contamination from animal reservoirs to the environment and foods1, on a broader geographic range2. Therefore, it can be expected that the incidence of infections with foodborne pathogens such as Escherichia coli O157:H7 and Salmonella will likely increase in regions experiencing significant climate change, providing enhanced opportunities for the survival, spread and increased virulence of pathogenic bacteria3,4.
There is a paucity of data in the scientific literature on the incidence of foodborne illness in the Arctic attributable to pathogenic bacteria. In the early 1990s, an outbreak linked to consumption of hamburger meat contaminated with E. coli O157:H7 occurred in Arviat, Nunavut5. Whale meat has been implicated as a vehicle for Salmonellosis in Tununat, Alaska6. No cases of human illness attributable to E. coli O157:H7 have been reported for the Nunavik region7, although at least 2 cases of gastroenteritis caused by pathogenic E. coli were reported in 2000 and 18 cases due to Salmonella in 19938. Furthermore, Nunavik has had the highest rate of gastroenteritis cases due to consumption of contaminated water in Quebec during the period 1991-20009,10. It may reasonably be expected that this region will be increasingly susceptible to changes in the prevalence, distribution and exposure to foodborne pathogenic bacteria due to modern lifestyle factors (eg increasing trend towards formation of higher density, permanently settled communities), eating habits (eg consumption of raw and dried game meats), infrastructure/geological factors (eg the disposal of human waste in open pits, and the melting permafrost which may increase runoffs towards river and lakes)8. Other factors making local populations more susceptible include the large number of visitors in communities such as Kuujjuaq (which serves as a transportation hub for the north), and the possible introduction of pathogenic bacteria from agricultural lands in the industrialized south through the agency of migratory birds, such as Canada geese11-13. In Nunavik, people regularly eat traditional foods in the following order of importance: caribou, arctic charr, goose, wild berries, ptarmigan, beluga blubber/misiraq, walrus, igunaq (fermented seal or walrus meat with blubber), scallop, seaweed and bear. Other animals consumed in lesser quantities include sculpins, northern pike, eider ducks, black scoter, northern pintail and murres14. These foods would likely be at risk of becoming contaminated by pathogenic bacteria present in the local environment, and thus serve as potential vehicles for human infection.
To face these new challenges, it will be necessary to gain a better understanding of the prevalence of key pathogens such as E. coli O157:H7 and Salmonella in arctic animal reservoirs and the environment, in order to predict their potential to spread in the human food chain. Furthermore, northern inhabitants will need to develop capability to monitor changes in their delicate environment, which is particularly vulnerable to the effects of climate change and demographic factors. An important element in this regard will be the ability to verify the safety of the local food supply. To this end, the present project encompassed two key objectives, to: (i) implement rapid, simple, and inexpensive test methods enabling the detection of the foodborne pathogens E. coli O157:H7 and Salmonella, suitable for transfer to a northern laboratory facility; and (ii) conduct a survey of the eastern Canadian Arctic in order to ascertain the prevalence of priority food pathogens in various traditional food animals.
This report summarizes the findings related to the study on the prevalence of E. coli O157:H7 and Salmonella in samples of traditional food animals from the Nunavik region of the eastern Canadian Arctic. In addition to obtaining as many samples from as great a variety of traditionally eaten wildlife as was logistically possible, the sampling strategy also involved visiting community freezers stockpiling locally harvested food animals, where samples were taken by swabbing various food contact surfaces to obtain a 'snapshot' of the general prevalence of the targeted pathogens in the local game meats trafficking through these facilities.
Methods
Selection of analytical methods for transfer to the Nunavik Research Centre laboratory
The first phase of this project focused on the identification of suitable analytical test methods enabling the detection of priority foodborne pathogens E. coli O157:H7 and Salmonella, for implementation at the Nunavik Research Centre (NRC) in Kuujjuaq, Nunavik, Canada. In order to establish basic food microbiology testing capability in such a remote location, where access to advanced technological support is limited, it was necessary to select rapid and simple methods for the preliminary screening of primary enrichment broth cultures for the presence of E. coli O157:H7 and Salmonella. It was intended that any positive samples identified using these screening methods would be shipped to the Ottawa Laboratory (Carling) of the Canadian Food Inspection Agency (CFIA) where further testing would be conducted to isolate, confirm, and characterize the organism.
There are a number of rapid test methods published in Health Canada's Compendium of Analytical Methods that have been validated and may serve as excellent choices for transfer to the NRC. The rapid test chosen for the presumptive qualitative detection of E. coli O157 (including H7) is the Merck Singlepath® E. coli O157 (MFLP-82)15. Singlepath® E. coli O157 is an immunological screening test based on the immune flow principle and is designed in such a way that time-consuming and personnel-intensive working steps are avoided. Singlepath® E. coli O157 has been validated and received AOAC approval (AOAC licence no. 010407) for use in raw ground beef and pasteurized whole milk. According to this license, the performance characteristics of the test kit matched the manufacturer's specifications, exhibiting a sensitivity of 97% and a specificity of 98.4%.
For the presumptive detection of Salmonella, the MFLP-75 'Procedure for the isolation of Salmonella species by the modified semi-solid Rappaport Vassiliadis (MSRV) method'16 was selected. This method is based on the ability of Salmonella species to grow and exhibit motility on MSRV medium after incubation at 42°C under adequate moisture conditions. Most strains of Salmonella species are able to migrate more than 20 mm from the inoculation site within 24-48 h. A collaborative study to validate motility enrichment on MSRV medium for the rapid detection of motile Salmonella in cocoa powder and chocolate concluded that the method had a sensitivity of 98.1% and a specificity of 100%, resulting in first action adoption of the method by AOAC International17. The MSRV medium has been used for the isolation of Salmonella from stool specimens18 and a variety of food products19 with high sensitivity and specificity.
Samples for analysis
Samples were collected by hunters from different communities of Nunavik (Table 1). The hunters were paid for each sample provided and received a training session on collection procedures prior to the sampling season. Tissue samples were kept frozen, brought to the community coordinator and sent to the NRC laboratory for analysis. Three sample types were collected: (i) meats from regionally harvested animals; (ii) intestinal contents (feces) of migratory geese; and (iii) environmental swabs of community freezers in Kuujjuaq and Tasiujaq. Samples collected are detailed (Tables 1,2). All animal derived samples were obtained from different individuals. These samples were sent frozen to the Ottawa Laboratory (Carling) for comprehensive microbiological analysis. Positive and negative controls were analyzed in parallel with each set of test samples: for E. coli O157:H7 analysis, the positive control consisted of an E. coli O157:H7 isolate (OLC-795) grown overnight in modified trypic soy broth with novobiocin (mTSB-n) (EMD Chemicals Inc, Gibbstown, NJ, USA), and the negative control consisted of mTSB-n broth devoid of bacteria; for Salmonella analysis, the positive control consisted of a Salmonella enterica ser. Berta isolate (OLC-759) grown overnight in buffered peptone water (BPW) (Becton, Dickinson & Co, Sparks, MD, USA), and the negative control consisted of BPW devoid of bacteria.
Table 1: Samples analyzed according to originating community and target animal
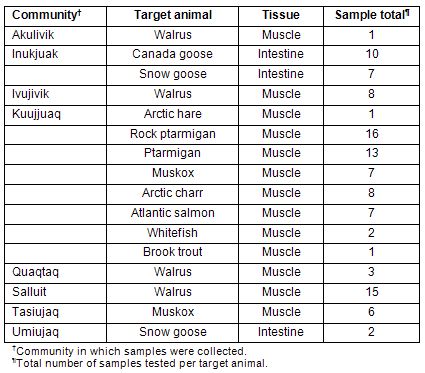
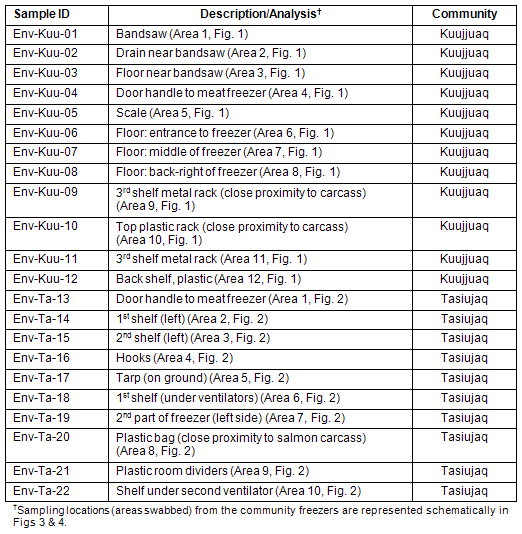
Table 2: Environmental samples collected from Kuujjuaq and Tasiujaq community freezers
General preparation, kits, and media
All media and reagents were prepared according to manufacturer's instructions prior to analysis. For each sample set, the required number of Singlepath® E. coli O157 test devices (EMD Chemicals Inc, Gibbstown, NJ, USA) were removed from 4°C storage and allowed to equilibrate to room temperature for at least 30 min in foil pouches prior to use. For sampling the surfaces in the community freezers, BPW containing 30% (v/v) glycerol (BioBasic Inc, Markham, Ontario, Canada) was prepared for use as the hydrating medium for the SpongeSicle? swabs (3M Canada, London, Ontario, Canada). Proficiency test (PT) samples were prepared by the Proficiency Testing Unit (PTU) of the Ottawa Laboratory (Carling) of the CFIA: Ground beef was lyophilized using the Tri-Philizer MP (FTS Systems Inc, Stone Ridge, NY, USA) followed by grinding in a Robot Coupe (Robot Coupe USA Inc, Ridgeland, MS, USA). Lyophilized ground beef was mixed with couscous in a 65:35 ratio to form the blank food matrix (BFM). The BFM was weighed in 11 g portions, placed in 30 mL-capacity Wheaton sample vials, and sterilized using irradiation. The PT samples were prepared as follows: (i) BFM samples were inoculated at a level of 103 CFU/g of meat with (a) E. coli O157:H7 (ATCC 35150), or (b) Salmonella enterica subsp. enterica ser. Heidelberg (ATCC 8326); and (ii) BFM samples devoid of inoculum were used as blanks.
Sample analysis
The PT samples were shipped to the NRC to assess the ability of laboratory staff to successfully identify the target pathogens in samples using the selected analytical methods described above. The inoculation status of the samples was not known at the time of analysis. Samples with the prefix 'PT-Salm' were to be analyzed for the presence of Salmonella, whereas samples marked 'PT-EcoO157' were analyzed for the presence of E. coli O157:H7. Each sample vial was aseptically opened and contents were transferred to a sterile filtered stomacher bag. The sample was rehydrated for 15 min in 100 mL of sterile diluent (mTSB-n for E. coli O157:H7 analysis or BPW for Salmonella analysis) and stomached for 30 s. Samples were incubated at 42°C (E. coli O157:H7) or 35°C (Salmonella) for 22-24 h. Enriched samples were then screened for the presence of E. coli O157 or Salmonella by subjecting to the Singlepath® E. coli O157 (MFLP-82)15 or MSRV (MFLP-75)16 methods, respectively. Details for each screening method are given below. Results were reported to the PTU of the Ottawa Laboratory (Carling).
Samples collected from the Nunavik region by NRC staff and community hunters were received in the frozen state and stored at -20°C. Prior to analysis, samples were removed from the freezer and allowed to thaw overnight at 4°C. Twenty-five grams of meat (whenever possible) (Table 1) were weighed in sterile filtered stomacher bags and stomached in 225 mL enrichment broth (or amount necessary to maintain a 1:10 dilution) (mTSB-n for detection of E. coli O157:H7 or BPW for detection of Salmonella). For environmental swabs (Table 2), sponges were swiped on different freezer surfaces and placed in 90 mL of the appropriate enrichment medium. A positive and a negative control were set up to run in parallel with the samples. Samples were incubated at 42°C (E. coli O157:H7) or 35°C (Salmonella) for 22-24 h. Enriched samples were then screened for the presence of E. coli O157 or Salmonella by subjecting to the Singlepath® E. coli O157 (MFLP-82)15 or MSRV (MFLP-75)16 methods, respectively. Details on each screening method are given below and schematic representations of the overall testing for E. coli O157:H7 and Salmonella are shown (Fig1 and Fig2, respectively).
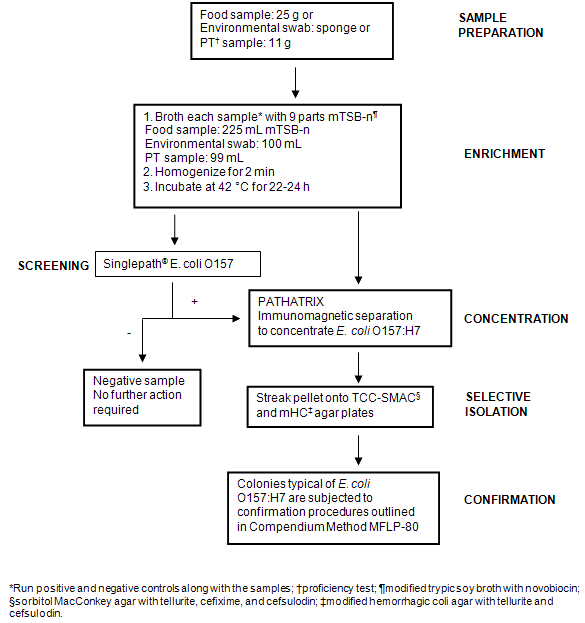
Figure 1: Flow diagram illustrating methodology used for the detection and isolation of Escherichia coli O157:H7 from samples.
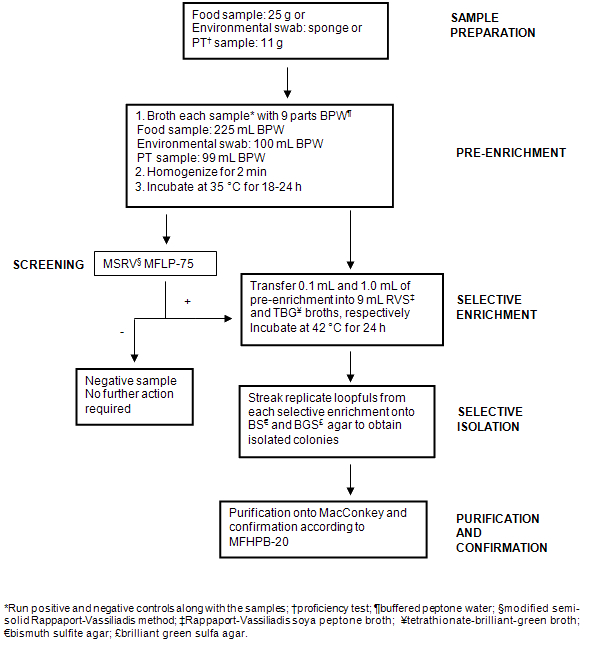
Figure 2: Flow diagram illustrating methodology used for the detection and isolation of Salmonella from samples.
Screening by Singlepath® Escherichia coli O157
Test devices were placed on a flat surface, labelled with appropriate sample identification, and utilized within 2 h from unpacking (as specified in the manufacturer's instructions). Using a pipettor and disposable filtered tips, 160 µL of the enrichment was dispensed into the circular sample port on the test device. Test results were recorded as positive or negative 20 min after applying the sample to the test device. A red line in the control zone (C) of the device within 20 min was regarded as valid; a sample was considered 'presumptively positive' if red lines appeared at both the test (T) and C zones within 20 min; a sample was considered negative if no red line appeared in the T zone after 20 min.
Screening by the semi-solid Rappaport Vassiliadis method
Portions of the pre-enrichment culture (0.1 mL) were inoculated on one side of an MSRV plate (Oxoid Ltd., Basingstoke, Hants, England), a few millimetres away from the edge. Positive and negative controls were inoculated onto separate plates. Inoculated MSRV plates were incubated in a high moisture incubator at 42°C for 24-72 h (due to the semi-solid state of the agar, plates were not inverted). Plates were examined after 24 h and the distance of any migration or swarming growth was measured (distance between the edge of the inoculation point and the farthest boundary of migration). If migration was not evident (less than 20 mm), plates were re-incubated and re-examined up to 72 h. If after 72 h the migration was still less than 20 mm, the MSRV result was considered negative for Salmonella. If the zone of migration was greater than 20 mm, the MSRV was considered 'presumptively positive' for Salmonella.
Environmental swabbing of community freezers
The community freezers of Kuujjuaq and Tasiujaq were tested for the presence of E. coli O157:H7 and Salmonella by swabbing the freezers in various areas (Fig3,4; Table 2). The Kuujjuaq community freezer was visited in May 2008. At the time of the visit, portions of caribou, fish, and fowl were observed and approximately 50% of the shelving units were stocked. The community freezer in Tasiujaq was visited in November 2008. The freezer contained portions of caribou, seal hides, fish, and fowl and was stocked at 25% capacity. For environmental swabbing, SpongeSicles? (pre-moistened in 10 mL BPW) were dipped in 10 mL of glycerol media (BPW containing 30% glycerol) and area of interest was swabbed horizontally and vertically in a zig-zag fashion. Sponges were deposited in sample bags and returned to the lab for analysis within 24 h after collection.
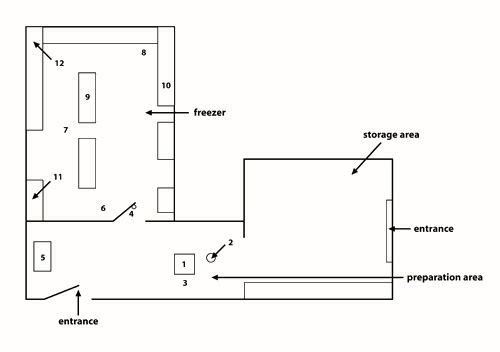
Figure 3: Schematic representation of the community freezer in Kuujjuaq. The community freezer in Kuujjuaq is organized into three separate compartments: (1) a general storage area; (2) a preparation area; and (3) a freezer.
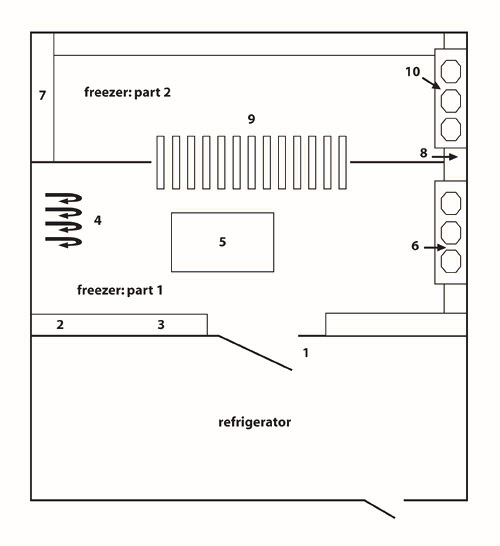
Figure 4: Schematic representation of the community freezer in Tasiujaq. The community freezer in Tasiujaq is organized into three compartments: (1) a refrigerator; (2) freezer #1; and (3) freezer #2.
Confirmation of suspected positive samples
Enriched samples identified as 'presumptively positive' for E. coli O157:H7 were subjected to immunomagnetic separation using the PATHATRIX® (Matrix Microscience, Golden, CO, USA) method according to manufacturer's instructions. Pellets recovered by the PATHATRIX® system were streaked onto modified sorbitol MacConkey agar with tellurite, cefixime, and cefsulodin and modified hemorrhagic coli agar with tellurite and cefsulodin, and then incubated at 42°C for 18-24 h. Plates were examined for E. coli O157:H7 colonies. Typical colonies were subjected to confirmation procedures outlined in Compendium method MFLP-80: 'Isolation of E. coli O157:H7 or NM in foods'20. Enriched samples were also confirmed by subjecting a portion of the enrichment to a PCR-based method according to MFLP-30 'The DuPont Qualicon Bax® system method for the detection of E. coli O157:H7 in raw ground beef and fruit juice'21.
For MSRV plates producing 'presumptively positive' results, a portion of swarming growth was inoculated on a MacConkey Agar plate (Becton, Dickinson & Co, Sparks, MD, USA) and incubated at 35°C for 18-24 h for isolation of Salmonella colonies. Typical colonies were subjected to confirmation procedures outlined in Compendium Method MFHPB-20: 'Isolation and Identification of Salmonella from Foods'22.
Results
Environmental swabbing of community freezers
The community freezers of Kuujjuaq and Tasiujaq were swabbed for the presence of E. coli O157:H7 and Salmonella. Environmental samples from the community freezer in Kuujjuaq were taken during the visit of Ottawa Laboratory (Carling) staff in May of 2008. At the time of sampling, SpongeSicles? pre-moistened in 10 mL of BPW were used for swabbing surfaces such as stainless steel shelving, floors and handling equipment. It was noted that the sponges had a tendency to freeze on contact with the freezer shelves, making it difficult to achieve an effective sampling of the area being swabbed. To remedy this, and for subsequent sampling exercises (ie sampling the Tasiujaq freezer), the possibility of moistening the swabs with BPW containing glycerol to prevent freezing was investigated. To determine the optimum glycerol content preventing freezing without impacting microbial growth, solutions of BPW containing various levels of glycerol (10-40% v/v) were assessed (Table 3). It was observed that BPW containing 30% (v/v) glycerol prevented freezing of the swabs on contact with a sub-zero metal surface, while not impairing the survival (and subsequent recovery) of bacteria. Therefore, SpongeSicle? swabs pre-moistened with this solution were used for swabbing the community freezer in Tasiujaq.
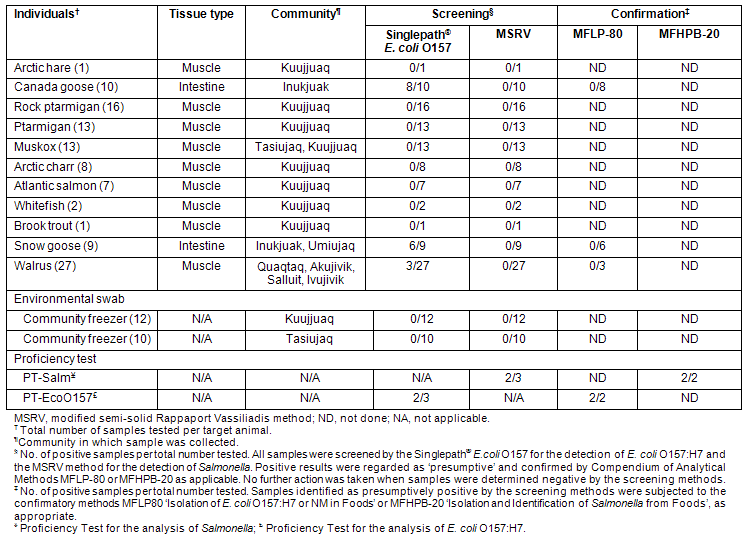
A total of 22 samples swabbed from different surfaces located within the two community freezers (Kuujjuaq and Tasiujaq) were tested for the presence of E. coli O157:H7 and Salmonella. None of the samples taken from these freezers were positive for the presence of either E. coli O157:H7 or Salmonella (Table 4), suggesting the absence of these pathogens (in viable form) from the surface residues of regionally harvested game meats, and by extension, from potential animal reservoirs associated with the general communities at the time of testing.
Table 3: Evaluation of glycerol media for use in conjunction with the 3M SpongeSicle? for the purpose of environmental swabbing of community freezers
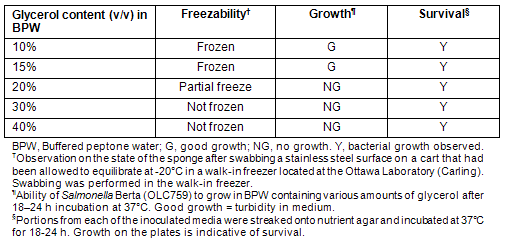
Table 4: Prevalence of Escherichia coli O157:H7 and Salmonella in samples collected in Nunavik
Prevalence of Escherichia coli O157:H7 and Salmonella in traditional animal foods
A total of 107 samples derived from 11 different species of animals were analyzed for the presence of E. coli O157:H7 and Salmonella in addition to the 22 freezer samples described above. These samples represent a broad cross-section of different types of game animals (terrestrial and marine mammals, fowl, and fish) (Table 1) harvested in Nunavik and consumed by the Indigenous populations of the eastern Canadian Arctic. The results obtained in the analysis of all samples in this study are summarized (Table 4). The results obtained with PT samples (ground beef inoculated with either E. coli O157:H7 or Salmonella) prepared by the CFIA PTU are included as controls.
While most samples of snow goose and Canada goose intestines, and some walrus meat, produced presumptive positive results for the presence of E. coli O157 in the screening phase, no E. coli O157:H7 colonies or BAX PCR-positive results (not shown) in enrichment cultures were obtained from these samples on subsequent attempts to confirm these results. None of the Nunavik samples produced presumptive results for Salmonella, nor was the presence of either pathogen confirmed in any of the samples tested. However, PT samples inoculated with these pathogens produced the expected results at both the screening and confirmation stages, confirming the validity of the test procedures. These results suggest that there was no evidence of prevalence for these pathogens among the different types of samples tested at the time of their analysis. It should be noted that all animal samples were received at the laboratory in the frozen state, and that even the freezer environmental samples were swabbed from sub-zero temperature surfaces. The impact of freezing on the viability of the target pathogens in such samples is unknown, nor was it systematically addressed in the present study, and may have been a factor in the failure to recover E. coli O157:H7 and salmonellae.
Discussion
The objectives of this project were two-fold, to: (i) implement appropriate test methods enabling the detection of the foodborne pathogens E. coli O157:H7 and Salmonella in foods suitable for transfer to establish on-site food microbiology testing capability in Nunavik; and (ii) conduct a survey of the eastern Canadian Arctic food animals to ascertain the prevalence of E. coli O157:H7 and Salmonella.
The establishment of on-site food microbiology testing capability at the NRC was achieved through the following means: (i) the purchase of basic microbiology equipment, including a bench-top autoclave, two incubators, a balance, a water bath, and pipettors, as well as all necessary reagents, media and disposables required to conduct microbiological analyses; (ii) hands-on training program conducted in the spring of 2008 to train NRC staff on basic enrichment culture techniques and on screening procedures selected for the detection of target foodborne pathogens in two phases: (a) NRC staff visited the Ottawa Laboratory (Carling) of the CFIA to receive basic training in food microbiology techniques and on Transportation of Dangerous Goods (for shipping of cultures and other materials associated with the subject analyses), and (b) CFIA staff visited the NRC laboratory in Kuujjuaq to help set up the equipment for food microbiology testing and to initiate sample collection and analysis (including sampling of the community freezers); and (iii) the effectiveness of training delivered to NRC staff was demonstrated through their ability to successfully identify the target pathogens in PT samples prepared and shipped by the PTU of the Ottawa Laboratory (Carling) to the NRC. For long-term implementation of food microbiology testing capability at the NRC site, it will be necessary to consider on-going verification of analytical proficiency through the periodic analysis of PT samples similar in nature to those utilized in the present study. The provision of such samples is highly feasible since there exist numerous commercial PT sample providers supplying food microbiology testing laboratories worldwide.
Due to its remote location and operating environment, the NRC laboratory is constantly faced with new challenges. Timelines for completion of analyses were impacted by the lack of human resources and facility problems, such as: (i) limited availability of laboratory space; (ii) logistical issues with air carriers for shipping materials and laboratory supplies to Kuujjuaq; and (iii) unavailability of distilled water on-site, hampering media preparation and the proper operation and maintenance of certain types of equipment such as the autoclave. Heavy workloads in diverse program areas at the NRC laboratory resulted in some difficulty to maintain a focus on the analysis of samples, therefore, it became necessary to ship some of the frozen samples to the Ottawa Laboratory (Carling) for analysis. It should be noted, however, that this project presented the first opportunity ever to conduct microbiological testing in Kuujjuaq, and local laboratory staff ultimately addressed the majority of the challenges. Nonetheless, the training of new local personnel for diagnostic testing will remain an issue, because high staff turnover is an inherent problem in Canadian northern communities. This is the main reason for the present approach of selecting simple tests (featuring ease of use and results interpretation) and limiting the involvement of the remote laboratory to the initial screening of samples for the purpose of identifying presumptive positives (which could be acted upon locally as a precautionary measure), with a more technically advanced laboratory conducting the more challenging aspects of pathogen recovery, identification and characterization.
No confirmed positives were obtained for either E. coli O157:H7 or Salmonella for any of the Nunavik samples examined. These results suggest two possibilities: (i) there is no evidence of the presence of E. coli O157:H7 and Salmonella in the Nunavik animal populations tested; or (ii) E. coli O157:H7 and Salmonella may have been present in low numbers, but were not detectable due to injury caused by sample freezing. It was noted that positive signals were obtained with the Singlepath® E. coli O157 test for most of the snow geese, Canada geese, and some walrus samples, but these could not be confirmed on subsequent analysis using cultural and PCR (BAX) techniques. These 'false positives' may be attributable to cross-reactions of the test reagents with the natural microflora in the samples. Alternatively, it is possible that E. coli O157 were present in these samples but were not recoverable using cultural techniques due to interference by the endogenous microflora. Therefore, the possibility that E. coli O157 is associated with the animals from which the samples were derived cannot be entirely discounted, and further studies may be warranted.
The effectiveness of analyzing samples collected from locally harvested game species in determining pathogen prevalence is limited by the total number and diversity of samples available. Furthermore, it was not possible for the present investigators to control precisely the nature and condition of samples provided by indigenous hunters and trappers, and it was necessary to proceed with the materials at hand. In the present study, 107 samples derived from 11 different species of animals, representing terrestrial and marine mammals, fowl, and fish, were analyzed.
Perhaps the most effective means of determining overall prevalence of the target pathogens in the region under study is the environmental sampling of the community freezers in the coastal communities of Nunavik during high-traffic hunting season. It is surmised that sampling such facilities provides a useful indicator of target pathogen prevalence since these facilities constitute a common focal point for the storage of approximately 30% of all locally harvested meat animals. Although there was no evidence of the pathogens targeted in this study, this may simply be an indication of the status of animal populations at a given point in time and seasonal cycle. Sampling a larger number of freezers from different communities throughout the eastern Canadian Arctic, and at different times of the year (following local hunting and trapping cycles) may yield a more accurate overall picture with regard to the prevalence of the target pathogens. A disadvantage of this approach is that any positive finding cannot be attributed to a point source or specific animal vehicle. Another limitation is the possibility that the sub-zero temperature environment may negatively impact on the successful recovery of viable bacteria. This limitation may be very difficult to overcome given the predominantly frigid character of arctic regions. Perhaps alternative analytical methods which do not rely on the recovery of viable bacteria, such as the use of polymerase chain reaction techniques to detect pathogen-specific DNA sequences, could be used as a means of inferring the presence of these pathogens. However, the inability to recover viable bacterial isolates with such an approach will not permit the definitive determination of the prevalence of these pathogens.
Conclusions
The results provide baseline data on the current prevalence of E. coli O157:H7 and Salmonella in traditional meats derived from game animals originating in the Nunavik region of the eastern Canadian Arctic. While these data indicate that E. coli O157:H7 and Salmonella were not prevalent in the samples collected from the Nunavik region, future sampling should be considered at periodic intervals to gauge the trends and risks to public health associated with changing demographics and the evolving natural environment.
In conclusion, the following points are offered:
- There was no evidence of the presence of Salmonella or E. coli O157:H7 in traditional meats derived from arctic animals harvested in the Nunavik region.
- The successful transfer of food microbiology test capability to a remote laboratory with basic facilities was demonstrated.
- The ability to conduct testing on-site will be essential in the timely implementation of appropriate measures to mitigate public health impacts (eg local food recall).
- The ability of the remote laboratory to provide high quality test results can be monitored on an on-going basis through the periodic provision of proficiency testing samples prepared in a reference facility.
Acknowledgements
The authors thank Stan Gagnon and Marie Claire Brassard from the Proficiency Testing Unit of the Ottawa Laboratory (Carling) for preparing proficiency test samples. The authors also express appreciation to Brygitte Gingras and Jessica Bosley from the Food Chemistry and Microbiology Section of the Ottawa Laboratory (Carling) for assisting with the BAX analyses. Gratitude is owed to Sandy Suppa, Wildlife Technician at the NRC of Makivik Corporation for his role in technical and coordination assistance. Thank you to Jeannie Gordon for providing technical assistance during sample analyses. The authors convey special thanks to all the coordinators and hunters from the Hunter, Fisher and Trappers Association and Hunter Support who collected field samples and shipped them to the NRC. The authors also acknowledge the participation of the ex-mayor Larry Watt from Kuujjuaq and mayor Peter Angnatuk from Tasiujaq for allowing them access to the community freezers. This project was partially funded by a Grant from the Government of Canada Program for International Polar Year.
References
1. D'Souza RM, Becker NG, Hall G, Moodie KB. Does ambient temperature affect foodborne disease? Epidemiology 2004; 15(1): 86-92.
2. Watson RT, McMichael AJ. Global climate change - the latest assessment: does global warming warrant a health warning? Global Change and Human Health 2001; 2(1): 64-75.
3. Kovats RS, Edwards SJ, Hajat S, Armstrong BG, Ebi KL, Menne B. The effect of temperature on food poisoning: a time-series analysis of salmonellosis in ten European countries. Epidemiology and Infection 2004; 132(3): 443-453.
4. Greer A, Ng V, Fisman D. Climate change and infectious diseases in North America: the road ahead. Canada Medical Association Journal 2008; 178(6): 715-722.
5. Orr P, Lorencz B, Brown R, Kielly R, Tan B, Holton D et al. An outbreak of diarrhea due to verotoxin-producing Escherichia coli in the Canadian Northwest Territories. Scandinavian Journal of Infectious Diseases 1994; 26(6): 675-684.
6. Bender TR, Jones TS, DeWitt WE, Kaplan GJ, Saslow AR, Nevius SE et al. Salmonellosis associated with whale meat in an Eskimo community. Serologic and bacteriologic methods as adjuncts to an epidemiologic investigation. American Journal of Epidemiology 1972; 96(2): 153-160.
7. La Direction des communications du ministère de la Santé et des Services sociaux. Surveillance des maladies à déclaration obligatoire au Québec - Rapport annuel 2002. Quebec: Ministère de la Santé et des Services sociaux du Québec (MSSS), 2005.
8. Martin D, Bélanger D, Gosselin P, Brazeau J, Furgal C, Déry S. Drinking water and potential threats to human health in Nunavik: adaptation strategies under climate change conditions. Arctic 2007; 60(2): 195-202.
9. Bustinza R, Levallois P. Étude de l'incidence des cas de gastro-entérites aiguë hospitalisés au Québec de 1991 à 2000 et évaluation de leur lien avec certains paramètres de qualité de l'eau potable. Quebec: Unité de recherche en santé publique, Centre Hospitalier Universitaire de Québec, 2003,
10. Anctil M. Les faits saillants de l'enquête. Enquête de santé auprès des Inuits du Nunavik 2004. Qanuippitaa? Comment allons-nous? Québec : Institut national de santé publique du Québec (INSPQ) et Régie régionale de la santé et des services sociaux du Nunavik (RRSSSN), 2008.
11. Cole D, David JV, Drum DE, Stallknecht DG, White MD, Ayers LS et al. Free-living Canada geese and antimicrobial resistance. Emerging Infectious Diseases 2005; 11(6): 935-938.
12. Hussein H, Abulreesh RG, Graham WS. Wild birds and human pathogens in the context of ringing and migration. Ringing and Migration 2007; 23(4): 193-200.
13. Sjölund M, Bonnedahl J, Hernandez J, Bengtsson S, Cederbrant G, Pinhassi J. et al. Dissemination of multidrug-resistant bacteria into the Arctic. Emerging Infectious Diseases 2008; 14(1): 70-72.
14. Blanchet C, Rochette L. Nutrition and food consumption among the Inuit of Nunavik. Qanuippitaa? How are we? Québec: Institut national de santé publique du Québec (INSPQ) and Nunavik Regional Board of Health and Social Services (NRBHSS), 2008.
15. Warburton D. Detection of E. coli O157 by the Merck Singlepath® E. coli O157 Kit (MFLP-82). In: Compendium of Analytical Methods, vol 3. Laboratory procedures of microbiological analysis of foods. (Online) 2007. Available: http://www.hc-sc.gc.ca/fn-an/res-rech/analy-meth/microbio/volume3/mflp-82-eng.php (Accessed 30 September 2009).
16. Poppe C, Mann ED, Shaw S, Warburton D, Sewell A. Procedure of the isolation of Salmonella species by the modified semi-solid Rappaport Vassiliadis (MSRV) method (MFLP-75). Compendium of Analytical Methods, vol 3. Laboratory procedures of microbiological analysis of foods. (Online) 2004. Available: http://www.hc-sc.gc.ca/fn-an/res-rech/analy-meth/microbio/volume3/mflp75-01-eng.php (Accessed 30 September 2009).
17. De Smedt J, Bolderdikjk R, Milas J. Salmonella detection in cocoa and chocolate by motility enrichment on modified semi-solid Rappaport-Vassiliadis medium: collaborative study. Journal of AOAC International 1994; 77(2): 365-373.
18. Dusch H, Altwegg M. Evaluation of five new plating media for isolation of Salmonella species. Journal of Clinical Microbiology 1995; 33(4): 802-804.
19. De Zutter L, De Smedt JM, Abrams R, Beckers H, Catteau M, de Borchgrave J et al. Collaborative study on the use of motility enrichment on modified semisolid Rappaport-Vassiliadis medium for the detection of Salmonella from foods. International Journal of Food Microbiology 1991; 13(1): 11-20.
20. Warburton D, Christensen D. Isolation of E. coli O157 in foods (MFLP-80). Compendium of Analytical Methods, vol 3. Laboratory procedures of microbiological analysis of foods. (Online) 2006. Available: http://www.hc-sc.gc.ca/fn-an/res-rech/analy-meth/microbio/volume3/mflp-80-eng.php (Accessed 30 September 2009).
21. Anon. The Dupont Qualicon Bax® system method for the detection of E. coli O157:H7 in raw beef and fruit juice (MFLP-30). Compendium of Analytical Methods, vol 3. Laboratory procedures of microbiological analysis of foods. (Online) 2003. Available: http://www.hc-sc.gc.ca/fn-an/res-rech/analy-meth/microbio/volume3/mflp30-eng.php (Accessed 30 September 2009).
22. D'Aoust J-Y, Purvis U. Isolation and identification of Salmonella from foods (MFHPB-20). Compendium of Analytical Methods, vol 2. Laboratory procedures of microbiological analysis of foods. (Online) 1998. Available: http://www.hc-sc.gc.ca/fn-an/res-rech/analymeth/microbio/volume2/mfhpb20-01-eng.php (Accessed 30 September 2009).


